


© Copyright 1995-2026 IMGT®, the international ImMunoGeneTics information system® | Terms of use | About us | Contact us | Citing IMGT
When several alleles are shown, the nucleotide mutations and amino acid
changes for a given codon are indicated in red letters. These polymorphic mutations are reported in
Tables of alleles.
Dashes indicate identical nucleotides. Dots indicate gaps according to the
IMGT unique numbering.
Blanks indicate partial sequences (blanks at the 5' and/or 3' end).
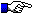 Germline genomic sequences are needed to confirm if the nucleotide substitutions result from somatic mutations or correspond to allelic polymorphisms.
Germline genomic sequences are needed to confirm if the nucleotide substitutions result from somatic mutations or correspond to allelic polymorphisms.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
C E Q L T Q P E S L T V R P G Q P L T
AJ274735,IGHV3S24*01, 09-43 TGT GAA CAG CTT ACT CAG CCC GAG TCT ... CTG ACT GTC CGA CCT GGT CAA CCT CTG ACC
AJ279392,IGHV3S24*02, 82 --- --- --- --- --- --- --- --- --- ... --- --- --- --- --- --- --- --- --- ---
___________________CDR1-IMGT____________________
21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
I R C Q V S Y S V S S Y Y T H
AJ274735,IGHV3S24*01, 09-43 ATC CGC TGT CAG GTC TCT TAT TCT GTT AGT AGC TAC TAC ... ... ... ... ... ACA CAT
AJ279392,IGHV3S24*02, 82 --- --- --- --- --- --- --- --- --- --- --- --- --- ... ... ... ... ... --- ---
_______________CDR2-
41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60
W I R Q P E G H A L E W I G R I V P G G
AJ274735,IGHV3S24*01, 09-43 TGG ATC AGA CAA CCT GAA GGA CAC GCA CTG GAA TGG ATT GGA CGG ATT GTT CCT GGG GGC
AJ279392,IGHV3S24*02, 82 --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- ---
IMGT________________
61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80
K T V K D S L S S K F S L D V
AJ274735,IGHV3S24*01, 09-43 AAA ... ... ... ... ACA GTG AAA GAT TCC CTT AGT ... AGT AAA TTC AGT CTT GAC GTT
V
AJ279392,IGHV3S24*02, 82 --- ... ... ... ... --- --- --- --- --- --- --- ... --- --- --- --- --- -T- ---
81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100
D G S S N T V T L Q G Q N L Q P G D S A
AJ274735,IGHV3S24*01, 09-43 GAC GGC TCC AGT AAC ACA GTA ACC CTG CAA GGG CAG AAC CTG CAG CCT GGA GAC TCA GCT
AJ279392,IGHV3S24*02, 82 --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- --- ---
CDR3-IMGT
101 102 103 104 105 106
V Y Y C A G
AJ274735,IGHV3S24*01, 09-43 GTG TAT TAT TGT GCC GGG #c
R
AJ279392,IGHV3S24*02, 82 --- --- --- --- --- A-A #c
#c: rearranged cDNA
This alignment of allele is automatically generated by Mehdi Yousfi alignment of allele tool.
Created: 21/01/2002
Author: Nathalie Bosc